Roots and shoots
Peter Kent
The quest to feed a hungry planet leads Julia Frugoli and her students straight to the root of the matter.
That smell, familiar but unexpected in a laboratory: just-cut grass, fresh fodder for livestock, the smell of chloroplasts breaking open.
“It smells like you just mowed your lawn,” says Julia Frugoli, a plant molecular biologist. She watches Ashley Crook, a Ph.D. graduate student in biochemistry and molecular biology who works with Frugoli, use a mortar and pestle to pummel plant leaves into powder.
If you’ve ever done this to herbs in your kitchen, you know that powdered parsley or basil isn’t the typical result.
Liquid nitrogen at –80 degrees centigrade makes the difference.
As soon as Crook pours liquid nitrogen over the leaves they flash freeze.
A few more minutes of freeze grinding and Crook collects the powdered cell parts and plant debris and puts them in a test tube containing a homogenized buffer with chemicals to sustain the proteins, DNA, and other research-worthy materials.
Voilà, crude lysate. It’s ready to be refined and put on a gel for analysis.
Crook does this three or four times a week with at least nine samples per session. The serrated leaves come from the small plants in trays under grow lamps by the window in a corner. They are mutant plants of an alfalfa, Medicago truncatula, the research model plant for Frugoli’s studies on how plants communicate, from shoots to roots and roots to shoots.
Challenge of the century
Plant molecular biologists, like Frugoli, along with many other scientists, are participants in the research challenge of the century: how to feed, clothe, and provide fuel for billions more of us without recklessly polluting the planet.
By 2050, forecasters estimate that the population will swell to 9.1 billion people, up from the current 6.1 billion.

Photo of Medicago truncatula, also known as barrel medic or barrel clover. The plant is native to the Mediterranean region and is often used in research. Photo courtesy of the Julia Frugoli lab.
Optimistic predictions state that we have barely adequate arable land, resources, and crop yields to make do—but these forecasts cannot accurately account for changes in climate and weather patterns during the next thirty-seven years. Plus, some studies do not address the environmental challenges of a rising global middle class that will want more meat on the dinner plate and more personal vehicles to drive. The pressures on intensive agriculture will increase water demand, dependence on fertilizer, particularly nitrogen, and the pesticides that cause pollution.
Many say that it will be plant scientists, plant breeders, and agrisystems engineers who will offer the greatest innovations for crop-yield advances. Basic research, such as Frugoli does, has the potential to contribute significantly.
Two topics are priorities in the Frugoli lab. One is figuring out how plants select whether to put more resources into what’s growing above ground or below it. The work could lead to controlling the process.
“If you could get more roots versus tops in, let’s say, carrots, or more tops than roots in lettuce, for example, you could adjust the allocation of energy in the plant,” Frugoli says. “If you understand how that decision is made, you can manipulate it.”
The second is the legume-rhizobia symbiotic relationship, which enables legumes to have a reliable source of usable nitrogen fixed by a bacteria living inside a root nodule. It’s a huge evolutionary advantage for legumes, Frugoli says, adding that it might be possible to activate this process in other plants that cannot make root nodules but could support the right bacteria. Frugoli’s lab works with an alfalfa, Medicago truncatula, to study how signals from root to shoot and shoot to root regulate root nodules, unique organelles where bacteria flourish and fix nitrogen.
Nitrogen is essential to life. It’s a basic ingredient in amino acids, found in proteins and nucleotides present in DNA and RNA. Plants need it for making chlorophyll molecules, key to photosynthesis. Animals get it by eating plants.
Nitrogen makes up about 80 percent of the atmosphere, but the gas itself is of little use to organisms unless it is “fixed.” In nature, microbes fix nitrogen and so does lightning, which has the power to break nitrogen molecules so that their atoms combine with oxygen to form nitrogen oxides. These oxides dissolve in rain, bathing plants and land with nitrates, a source of nitrogen plants can use. To make nitrogen fertilizer, industries break nitrogen molecules using a high-temperature, high-pressure process that yields ammonium.
Legumes, the third largest plant family, have evolved another way to get nitrogen. Alfalfa, beans, clover, lentils, peanuts, soybeans, and vetch are some of the legumes able to host bacteria that can take nitrogen from the air and convert it into ammonium usable by the plant. Civilization has benefitted from the symbiotic relationship between legumes and rhizobia for thousands of years. Farmers grow legumes not only for their protein value— legumes make up about a third of the world food supply—but also as “green manure.” Left to die in fields, legumes release their nitrogen into the soil, where it becomes nitrates, making the nitrogen useful to other plants.
Each legume has an exclusive rhizobium. Alfalfa’s is Sinorhizobium meliloti. In a complicated sequence of steps, bacteria in the nodules fix nitrogen that is converted biochemically into ammonium, which the plant moves up the shoot to make chlorophyll, proteins, and seeds.
For the plant, establishing and maintaining root nodules takes significant energy. The plant does it out of necessity, not convenience.
Signs of starvation
When a legume lacks nitrogen, growth slows or stops and the leaves turn yellow. If the condition is sufficiently dire, it can activate a “stringent response,” which usually indicates starvation. Then the legume has to make a decision. Root nodulation puts a strain on the plant, enough so that legumes resist starting up the process. The plant’s regulation of nodulation is a “negative feedback inhibition system,” as Tessema Kassaw wrote last year in Plant Methods. Kassaw, also a Frugoli lab grad student, completed his Ph.D. in December.

Julia Frugoli talks with Tessema Kassaw, a postdoctoral researcher (right), about the best way to set up a root graft to study how roots signal to other roots through the shoot of the plant. Listening is Elise Schnabel, a research technician. Photo by Craig Mahaffey.
“It’s like having a roommate,” Frugoli says about the plant’s decision. “If you can’t pay the rent, you need a roommate, but if you can pay, it might be nicer to be by yourself and not have to share. That’s kind of how the plant looks at the interaction. It has to decide, ‘Do I need nitrogen or not?’”
The result is observable: Either the plant makes root nodules or it doesn’t. But nodulation is a complex process, and researchers have a long way to go to explain it.
Like genetics researchers everywhere, Frugoli breaks things to study them. She uses mutagenesis, treating a plant with radiation or chemicals that jumble the DNA code, creating plants with atypical characteristics—mutant clones. She and her graduate students look for phenotypes, the observable traits of gene expression.
“Once we have plants that have the phenotype we are looking for, we go through the slow process of what’s called mapping, trying to determine what the gene we messed up is,” Frugoli says. “Once we find it, that’s only the beginning, because we have to figure out what that molecule does and how it does it. Sometimes the traits are not observable. The signals and the changes that come about are deep in the structure or system.”

Julia Frugoli: “If you could get more roots versus tops in, let’s say, carrots, or more tops than roots in lettuce, for example, you could adjust the allocation of energy in the plant." Photo by Craig Mahaffey.
M. truncatula improves the chances of finding the right molecule. It is a diploid form of alfalfa, meaning it has only two copies of each of its 40,000 or so genes. Other alfalfa varieties, such as the one that is used as cattle fodder, have four copies of each gene. The possible combinations and the search among them becomes a lot more time consuming and harder.
Crook and Kassaw have projects that fluorescently tag proteins so that they and Frugoli can look inside the plant while it’s alive.
“We see where the protein is, what it’s doing, and where it’s going—the pictures of glowing plant roots are kind of cool.”
In the lab, Crook works with a mutant of a shoot-to-root signaling gene that transmits a message from the leaves of alfalfa to the roots to grow root nodules. Its well-suited name is SUNN, Super Numeric Nodulator. The mutant disrupts the gene expression that stops nodule making. Mutant SUNN causes the plant to make too many nodules, squandering energy.
Another gene the lab studies is RDN1, Root Determined Nodulator. It goes from root to shoot, signaling when to stop nodule making. When RDN1 is missing, the plant also makes too many nodules, but for a different reason.
SUNN is a protein kinase, while RDN1 is a protein that remains uncharacterized, though it occurs in most plants. Protein kinases are enzymes—biochemical catalysts—that modify other proteins controlling growth and development. Important and ubiquitous, “conserved” kinases can be found in animal, bacteria, and plant genomes. This suggests that, as organisms have evolved and diversified, kinases were useful substances that served many different species. Kinases are particularly important in controlling activities in cells and cell pathways, especially signal transduction, which is a kind of communication.
How a plant makes decisions
All organisms send signals to grow and reproduce, to defend against disease and deal with environmental stresses. Signals can go short distances—inside cells, cell to cell—and long distances— from shoot to root, brain to big toe.
But plants rely on signals in a way animals do not. Plants are sessile, rooted in place. Coping with droughts, poor soil, temperature swings, diseases and insects, plants face a stark reality of working with what’s available. Make do, or die.
“Anytime something happens to plant, it has to change its gene expression essentially to respond,” Frugoli says. “It has to make a molecule that does something to change the way it grows. If a plant wilts, if it gets too much light or not enough light, if it gets hot, if gets cold—even if something bruises it or chews on it— all these things require a response.”
Something happens in a cell to trigger a molecule that begins a signaling process that goes through the plant’s xylem or phloem, from cell to cell.
“Every cell must make a decision based on that signal and passes the signal along to wherever it has to go,” Frugoli says. “So it’s either one cell at a time or it’s through the vasculature of the plant.”
Signaling is not like a light switch; it’s more like a marching band, each member on the move to a location, playing his or her part, creating music in formation. Out of step or off tune, missed notes, the intended result fails to happen.
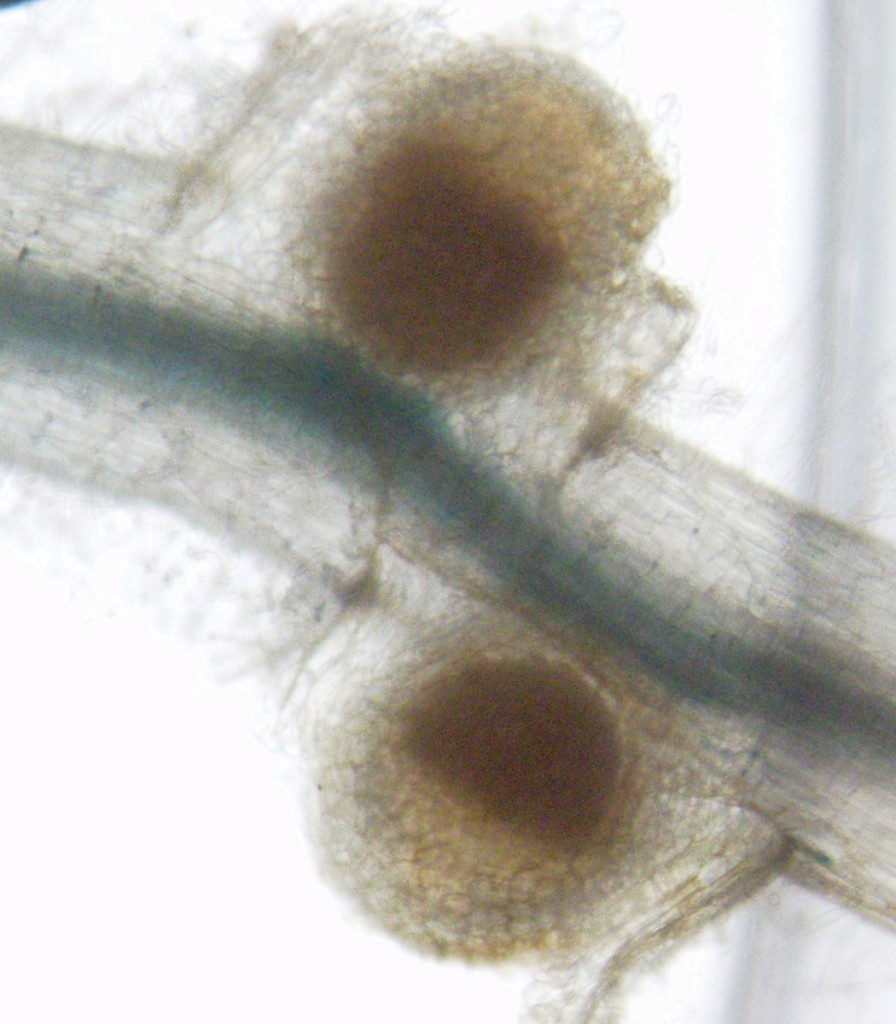
Nodules, like these on the roots of Medicago truncatula, are where bacteria take nitrogen from the air and convert it into ammonium usable by the plant. Photo courtesy of the Julia Frugoli lab.
“In a lot of these communications from roots to shoots, if there’s misstep, there’s a very subtle phenotype, a very subtle thing you can see,” Frugoli says. “Maybe the roots are a little bit shorter, or they are not as fuzzy.”
Legume roots turn out to be sensitive organs. They grow outward more than downward, seeking fast food, as do most annual plants faced with a seasonal deadline to flourish and reproduce. The bigger, thicker root parts mainly transport food up the stem. It’s the root tip where the action is.
All roots have molecular receptors and membranes on the outside of the root tip to detect chemical concentrations in the soil. Root tips can forage, acting like Geiger counters. When enough receptors measure an attractive level of a nutrient—nitrogen, for example—on one side of the root tip, the root will turn gradually toward the nitrogen if the amount and proximity is worth the energy to make the move. They can also forage for water.
Legume roots can do more. They can send signals out to invite the rhizobia to come and infest a root hair growing in a special crooked shape that leads to infection threads, allowing the bacteria to move across the outer cell layers and colonize the root nodule.
“Every gene we have discovered so far that is involved in nodulation, whether it’s regulating number of nodules or allowing nodulation to happen, isn’t exclusive to legumes,” Frugoli says. They occur in other plants that don’t make nodules. That doesn’t mean the other plants don’t have the right genes; we don’t know how to turn them on and off in a way will allow the rhizobia to interact with the plants.
Nodulation and nitrogen fixation have been a holy grail for plant geneticists and breeders. The dream of nodulation in corn, a nitrogen-intensive crop, has been around for nearly half a century. If corn could make its own nitrogen instead of relying heavily on nitrogen fertilizer, “we would be golden,” Frugoli says.
The hope was that it would take only a few added genes to create a super corn. It is, of course, more complicated, and researchers have yet to identify all the genes involved or to know with certainty how nodulation is regulated in plants.
Frugoli is a fan of another crop being the first to nodulate. “I would choose rice, because there are some things rice can do that are very close to nodulation, but not exactly.”
In the lab, Frugoli’s researchers have taken a rice gene and inserted it into an alfalfa mutant they created, and the combination behaves normally. “That means the rice gene is restoring the trait. We call it ‘rescuing the phenotype,’” Frugoli says. “When we put it in the plant in the right place and at the right time, it works. That suggests to us that it may be a problem of timing in rice.”
While rice looks to be a better fit than corn, nodulation in rice won’t happen today or tomorrow and may never come to be. But the basic knowledge Frugoli contributes may eventually help to grow more food or to pollute less by minimizing fertilizer use. And for a world facing the prospect of hunger, that kind of work sends all the right signals.
Julia Frugoli is an associate professor of genetics and biochemistry in the College of Agriculture, Forestry, and Life Sciences. The National Science Foundation supports Frugoli’s research. In 2012 she received a $600,000 four-year grant toward developing a model of the pathways plants use to pass messages back and forth from cell to cell. Peter Kent is a news editor and writer in Clemson’s Public Service Activities.


